As a final test of the release version of the MaSuRCA 3.2.2 assembler, I created an assembly of the human NA12878 data set from ~30x coverage of Oxford Nanopore data (rel3, https://github.com/nanopore-
The hybrid assembly took about 50,000 CPU-hours on my AMD Opteron 6000-series 400-core cluster. The assembly had the following quantitative statistics(G=3Gb):
Update 05/23/2017: Just for the sake of comparison here are the stats of the MaSuRCA assembly that used only the Illumina data:
Addition of the nanopore data makes huge difference in the assembly contiguity.
Compared to the available Canu Nanopore-only assembly of the rel3 data, The MaSuRCA assembly has 37% bigger N50 contig size, and has about 9% more sequence:
https://genomeinformatics.
The MaSuRCA assembly aligns to the GRCh38.p10 human reference (this is not the same human) at average 99.73% identity for 1-to-1 alignments.
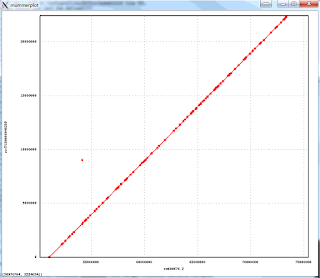
The 1-to-1 alignments to Illumina-only assembly of the NA12878 have an average identity of 99.96%, implying consensus error rate of 4 errors per 10,000 bases. Below is an alignment plot created by mummerplot of the biggest scaffold in the assembly (22.4Mb) aligning to the corresponding GRCh38 chromosome, that shows excellent large scale agreement:
The assembly is available here:
ftp://ftp.genome.umd.edu/pub/NA12878/assembly.fa
I am now working on validation of the structure of the assembly and classification of consensus errors.
Data used. For this assembly I used the publicly available Oxford Nanopore MinION data set rel3:
https://github.com/nanopore-
and about 100x coverage by Illumina 2x150b HiSeq 2500 Paired End reads from the Genome In A Bottle project:
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NIST_NA12878_HG001_HiSeq_300x .
I used the following Illumina runs from that FTP site:
131219_D00360_005_BH814YADXX
131219_D00360_005_AN81VLADXX
140115_D00360_0009_AH8962ADXX
140207_D00360_0013_AH8G92ADXX
The hybrid assembly took about 50,000 CPU-hours on my AMD Opteron 6000-series 400-core cluster. The assembly had the following quantitative statistics(G=3Gb):
Sequence in scaffolds: 2.88Gb
NG50 scaffold size: 5.04Mb
NG50 contig size: 4.06Mb
Update 05/23/2017: Just for the sake of comparison here are the stats of the MaSuRCA assembly that used only the Illumina data:
Sequence in scaffolds: 2.81Gb
NG50 scaffold size: 0.065Mb
NG50 contig size: 0.068Mb
Addition of the nanopore data makes huge difference in the assembly contiguity.
Compared to the available Canu Nanopore-only assembly of the rel3 data, The MaSuRCA assembly has 37% bigger N50 contig size, and has about 9% more sequence:
https://genomeinformatics.
The MaSuRCA assembly aligns to the GRCh38.p10 human reference (this is not the same human) at average 99.73% identity for 1-to-1 alignments.
The 1-to-1 alignments to Illumina-only assembly of the NA12878 have an average identity of 99.96%, implying consensus error rate of 4 errors per 10,000 bases. Below is an alignment plot created by mummerplot of the biggest scaffold in the assembly (22.4Mb) aligning to the corresponding GRCh38 chromosome, that shows excellent large scale agreement:
The assembly is available here:
ftp://ftp.genome.umd.edu/pub/NA12878/assembly.fa
I am now working on validation of the structure of the assembly and classification of consensus errors.
Data used. For this assembly I used the publicly available Oxford Nanopore MinION data set rel3:
https://github.com/nanopore-
and about 100x coverage by Illumina 2x150b HiSeq 2500 Paired End reads from the Genome In A Bottle project:
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NIST_NA12878_HG001_HiSeq_300x .
I used the following Illumina runs from that FTP site:
131219_D00360_005_BH814YADXX
131219_D00360_005_AN81VLADXX
140115_D00360_0009_AH8962ADXX
140207_D00360_0013_AH8G92ADXX
This is great! I wondered if you had used yet the additional files from the rel4 release which contains the very long read dataset?
ReplyDeleteI used all data from rel3 release. I did not use rel 4 data for this assembly, I am re-running the assembly with rel4 data now.
ReplyDeleteHuman NA12878 hybrid 30x MinION+100x Illumina assembly by MaSuRCA 3.2.2 ... MaSuRCA 3.2.2 assembler, I created an assembly of the human NA12878 ...
ReplyDeletegoldenslot
สูตรบาคาร่า